...
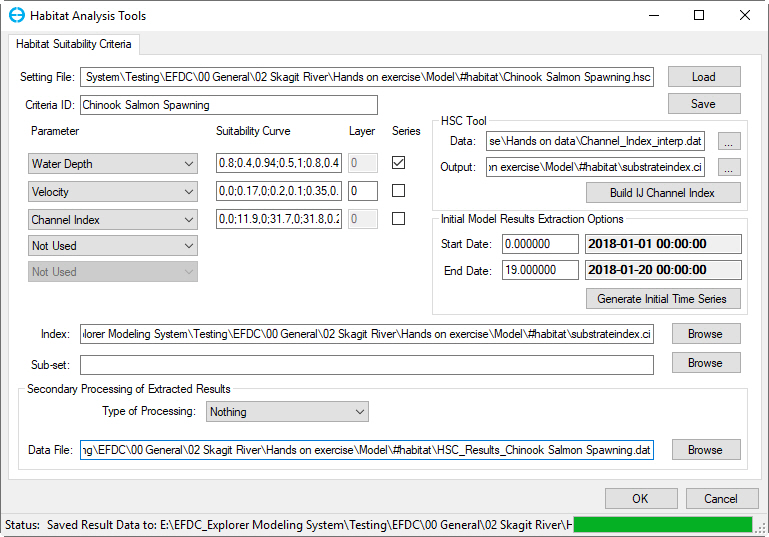
The interface for the IFIM approach to configuring habitat suitability criteria in EE is shown in Figure 1. When saving and loading the configuration files for habitat suitability criteria, EE uses “.HSC” as the extension.file extension and it's directory path will be displayed in Setting File box
The Criteria ID will be used to manage the different species. The Criteria ID is appended to the file name “HSC_Results” of the data file extracted when generating an initial time series. For the example shown in Figure 1 the extracted file will be called “HSC_Results_Walleye – Open WaterChinook Salmon Spawning.dat”.
| Anchor | ||||
|---|---|---|---|---|
|
Figure 1 Model Analysis: Habitat Suitability Criteria form.
...
After defining the SI curves the user should select the layer range as described in the Critical Limits section. It is required to select the checkbox to assign which series is to be plotted, and the start and end date of extraction from the model, before selecting Generate Initial Time Series.
Anchor Figure 2 Figure 2
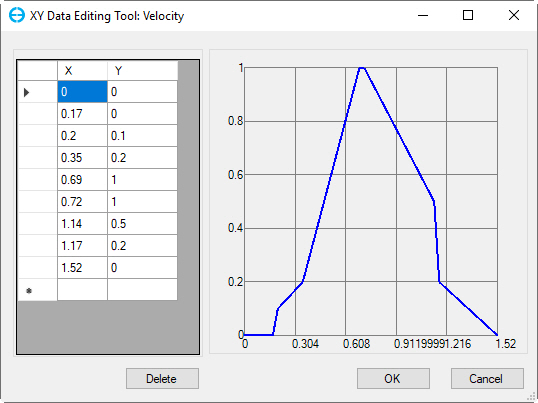
Figure 2 Editing tool for suitability index curves.
...
If the user doesn’t have a channel index file already, they should first build the channel index with the Build IJ Index button . Selecting this will open the tool shown under HSC Tool frame in Figure 31. In this form the frame, user should browse to import a channel index file from another tool such as River2D (.CHI) and import this file. River2D such as Channel_index_interp.dat to Input form. Channel_index_interp.dat file format is: “X, Y, Elevation, Substrate Code”, where X and Y should be in UTM. However, EE will prompt the user for the column containing the channel index so other formats can also be imported. Alternatively, the user can generate this externally in a spreadsheet. EE will interpolate the channel index to the I, J grid. The user selects the name of the output file, which will be saved in the EE file format with extension .CI and of the type: I, J, Channel Index.
...
.
Once the user has a channel index file already created they should browse to it in the it will be automatically browsed to Index box. As described in the critical limits section, it is also possible to generate the time series based on only a sub-set of the model domain by browsing to and selecting a polyline file with the Sub-Set box. If no file is selected in the Index option EE will effectively ignore this option by assuming the index is 1.0 everywhere.
...
To create time series EE will extract all of the cell information and save it in a post processing file in the #habitat folder with a name based on the Criteria ID . and it's directory path will be displayed in Data File box. This allows users to reload and review the results at anytime without generating time series. If the criteria ID is “Salmon01” “Chinook Salmon Spawning” then the file will be called HSC_Results_Salmon01Chinook Salmon Spawning.dat. This file records the composite time, salinity, net weighted criteria (weighted average 0 and 1 of all the cells that meet the bathymetry criteria), area, and volume.
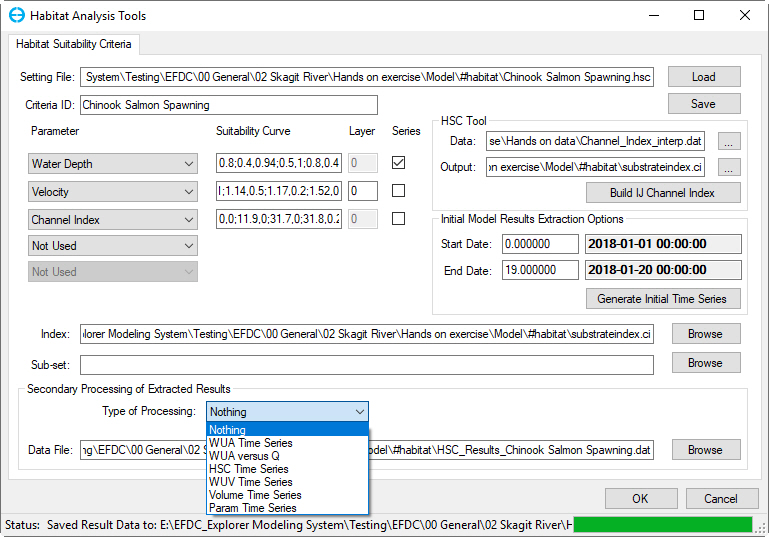
Once a time series has been generated, and the post-processing files have been extracted, a new frame is displayed, Secondary Processing of Extracted Results as shown in Figure 43.
Anchor Figure 43 Figure 43
Figure 4 3 Suitability Criteria: Secondary processing.
...
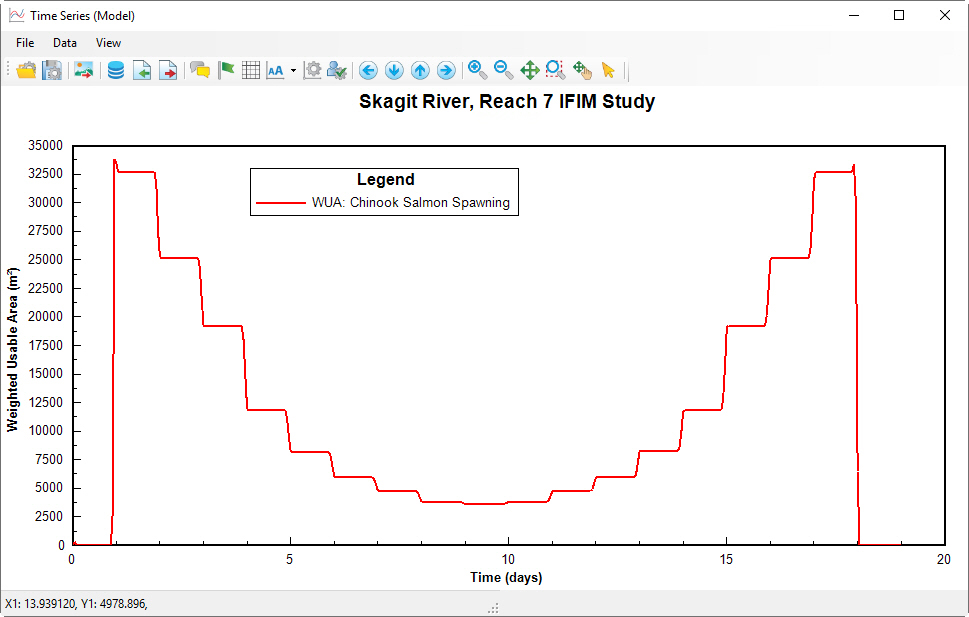
After selecting one of these options EE will display a button to the right of the menu. Selecting this button will display the time series such as that for the WUA time series shown in Figure 54. If the user has extracted the results previously in this session, and then wants to view another secondary processing type, EE will prompt whether to use the data in memory, or whether to extract the data again.
| Anchor | ||||
|---|---|---|---|---|
|
Figure 5 4 Weighted Usable Area time series.
View Habitat Suitability Index in 2DH View
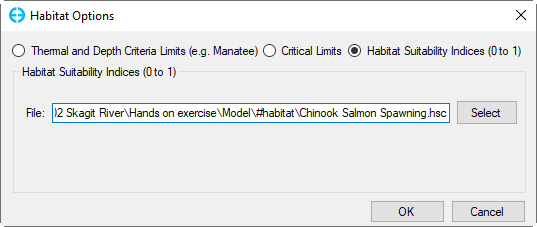
Once a time series has been generated, user can go to 2DH View window to view the habitat suitability index of each cell change by the time. From 2DH View window, users press keystroke Ctrl + H to open Habitat Options form as shown in Figure 5. From this form, select Habitat Suitability Indices option then click on Select button to browse to *.HSC file.
| Anchor | ||||
|---|---|---|---|---|
|
Figure 5 Habitat Options form.
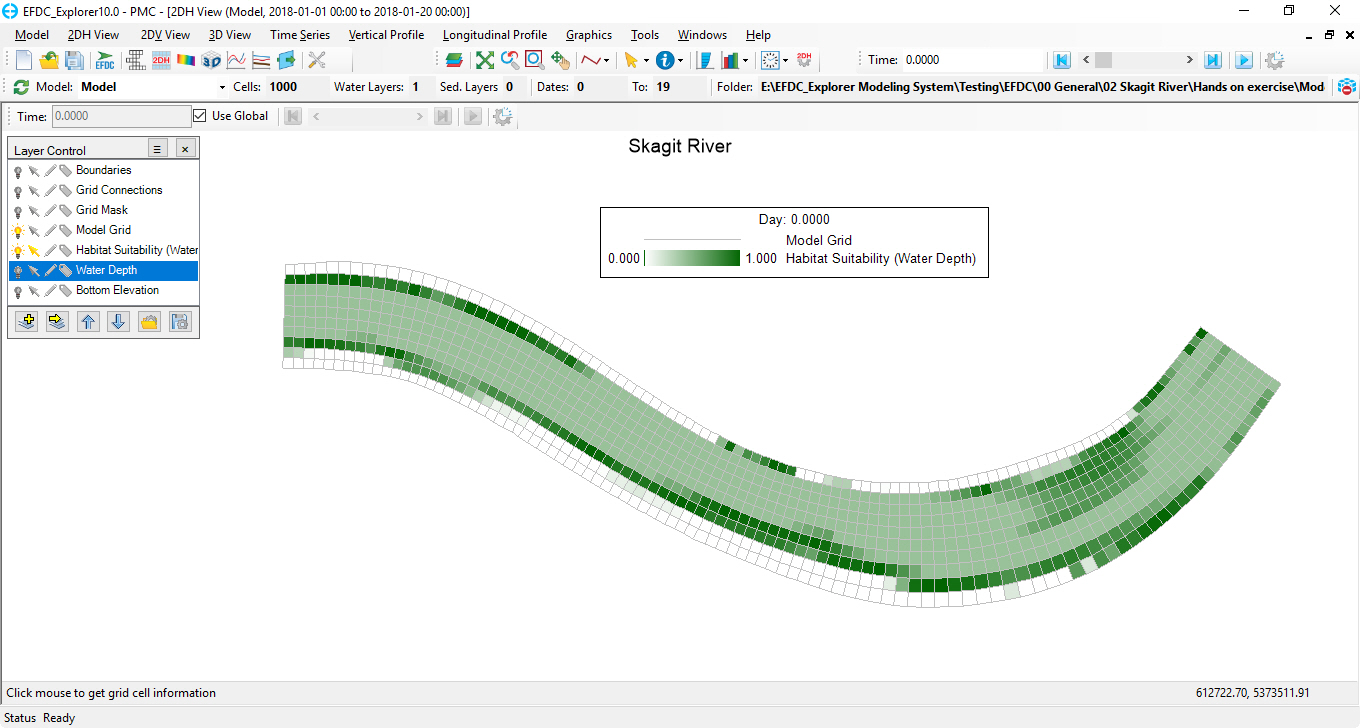
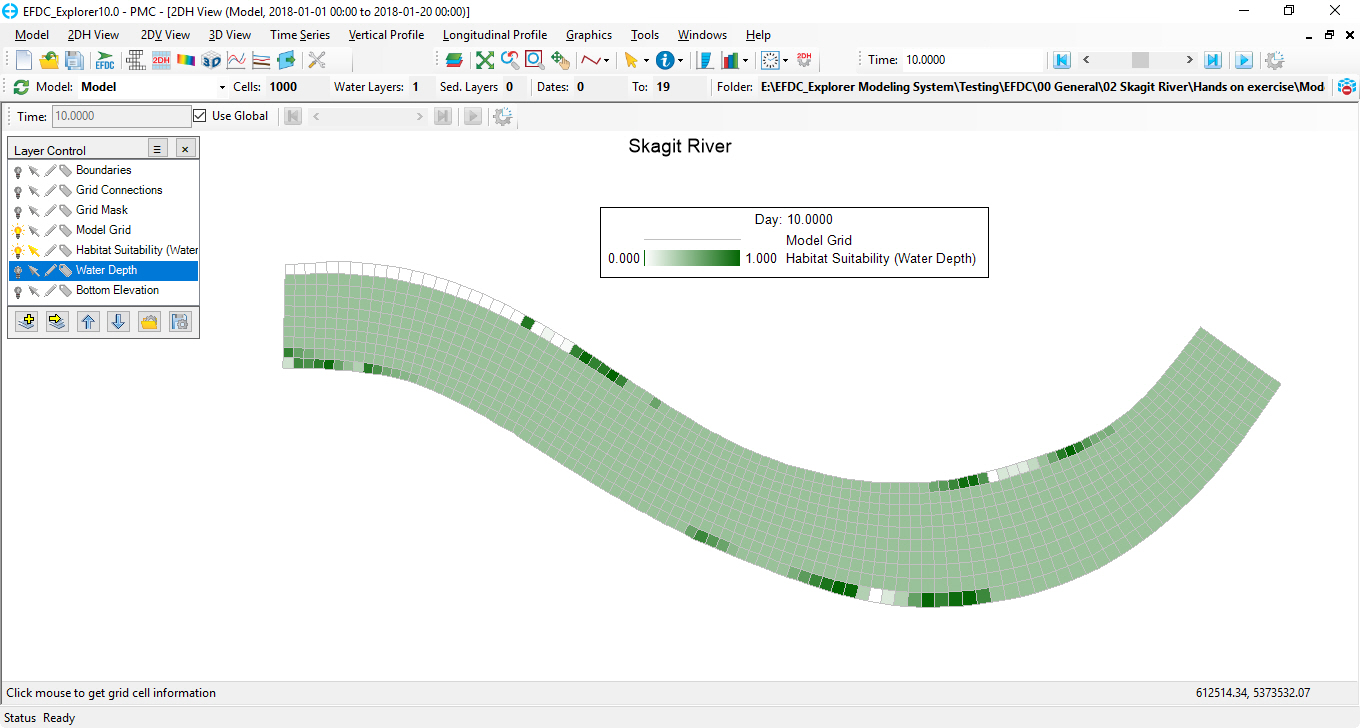
Habitat Suitability index of the interested species will be displayed at each time step as shown in Figure 6 and Figure 7 below.
| Anchor | ||||
|---|---|---|---|---|
|
Figure 6 HSC results at time step 0 in 2DH View.
| Anchor | ||||
|---|---|---|---|---|
|
Figure 7 HSC results at time step 10 in 2DH View.